-
Sep 13, 2020
Populating a Google Sheet with Goodreads data
Here’s an example of how you can use the Goodreads API with Google Sheets and Apps Script to automatically populate a sheet with book data based only on the book’s ISBN.
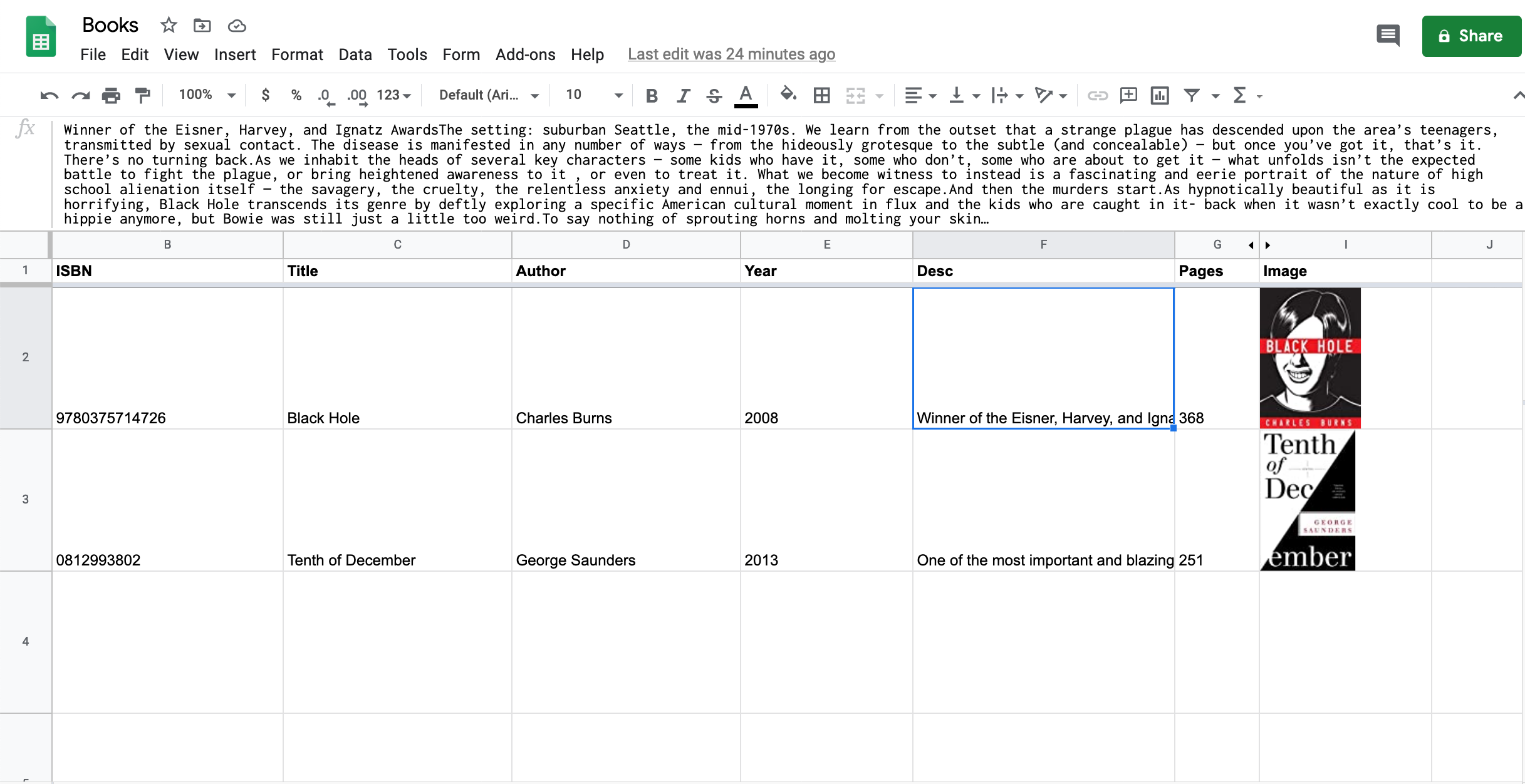
const API_KEY = "REPLACE_ME"; // paths to fields to extract from xml // e.g., ["authors", "author", "name"] maps to "Charles Dickens" in this XML: // <authors> // <author> // <name>Charles Dickens</name> // </author> // </authors> const FIELD_PATHS = [ ["title"], ["authors", "author", "name"], ["publication_year"], ["description"], ["num_pages"], ["image_url"], // to display an image from this url, create a formula cell // with `=IMAGE(X)`, where X is the cell containing this url ]; function bookDataFromISBN(isbn) { // isbn field must be text instead of number so that leading 0s are retained if (isbn == "") { return; } const url = `https://www.goodreads.com/book/isbn/${isbn}?format=xml&key=${API_KEY}`; const xml = UrlFetchApp.fetch(url).getContentText(); const document = XmlService.parse(xml); const root = document.getRootElement(); const book = root.getChildren("book")[0]; return [FIELD_PATHS.map(fieldPath => { return fieldPath.reduce((obj, pathPart) => { return obj.getChild(pathPart); }, book).getText().replace( /(<([^>]+)>)/ig, ''); })]; } -
Sep 12, 2020
A simple, templatized web page with Google Sheets
In the past, I’ve used Google Sheets to display data from the Google Places API and to keep score in Jeopardy!.
This post further explores the Google Sheets Apps Script integration. It shows how you can define a simple templatized web page in a sheet. You may find this useful if you’re building a small prototype, need to quickly put a customizable page on the web, or if you, like me, just like playing with Google Sheets. A web page built in this way is free to host, and is very easy to deploy and customize. That said, I wouldn’t recommend this setup for maintaining a production web application because it’s brittle and non-standard, and, since 2017, Google places a security header at the top of the web page (though the header can be hidden with a browser extension).
Google Apps Script is a powerful low-code solution for building web-apps, and it can do much more than what’s covered in my blog post here. There are other tutorials online about how to create web apps with Google Apps Script. This post instead focuses on using a minimal, generic script, and moving web page customization into the sheet itself. It does this by having the sheet store the dynamic HTML template, and cells that are fed into the template are designated within the sheet.
Data
The toy web page we’re about to build needs some data. Let’s say we’re making a company’s lunch menu that will be displayed on monitors outside the lunch room. First, navigate to https://sheet.new to create a new Google Sheet. Maybe we’ll show the following dynamic information:
- day of the week
- menu theme
- appetizers
- entrees
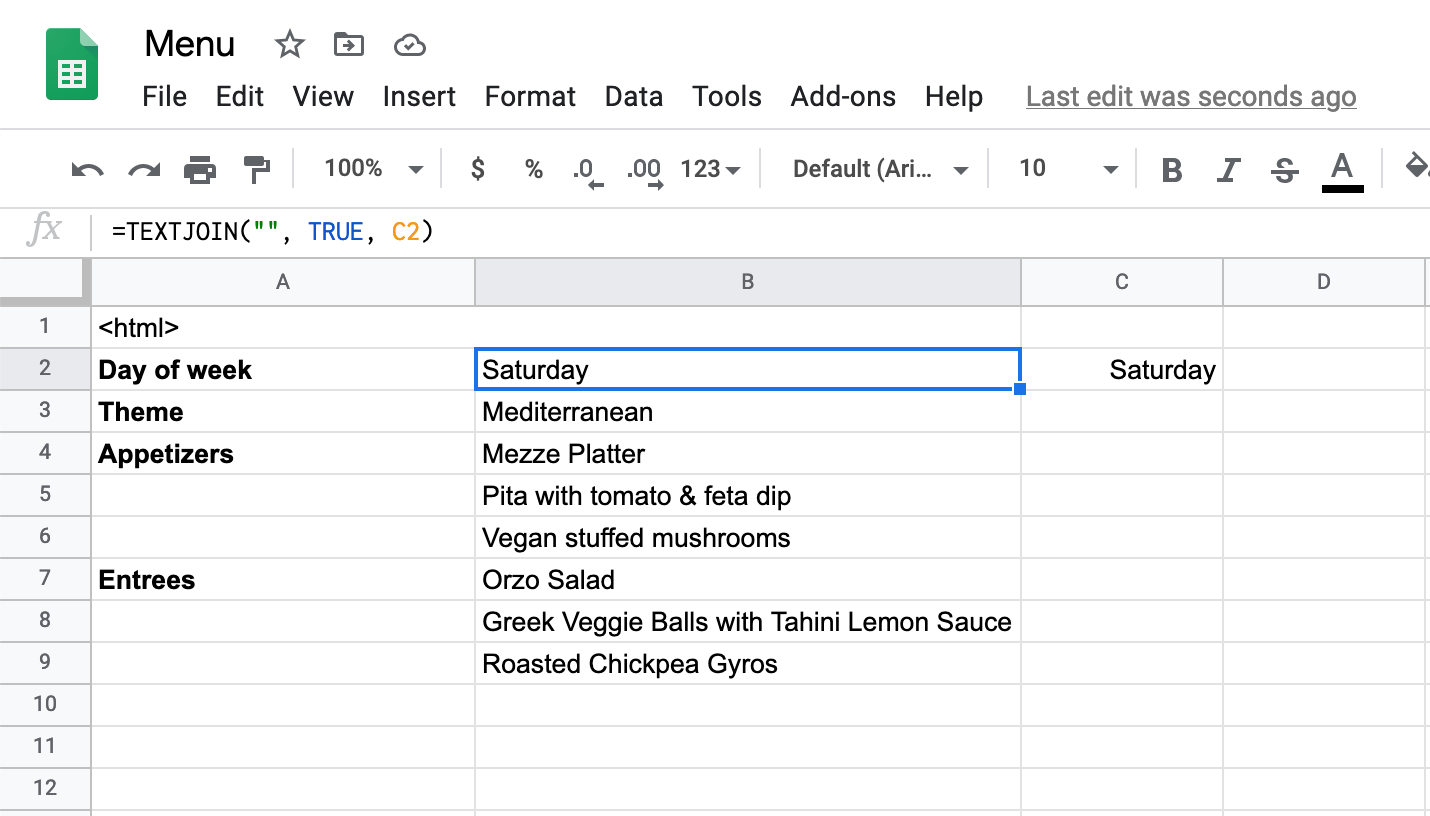
A couple notes… Ignore cell A1 for now, that’s where we’ll put the steak sauce, err I mean that’s where we’ll put the HTML template. Note that the cell in B2 contains text data, not an actual date. This is necessary because the script we’ll add later doesn’t know how to format the raw data it receives, so we’ll do that formatting directly in the sheet. You can make a dynamic day of week field by using the formula
=TODAY()and then changing the date format to only display the day of the week.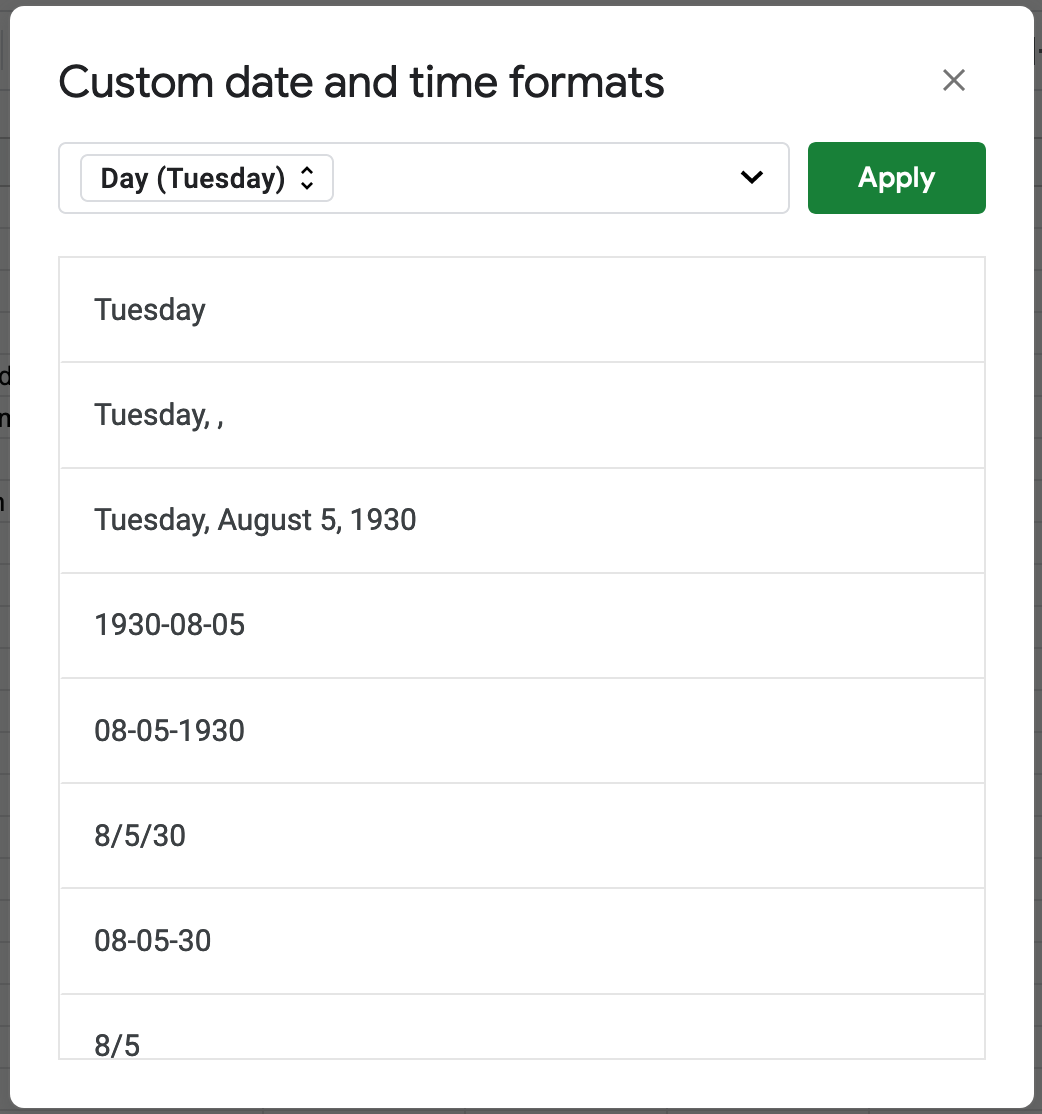
So note that, using the sheet itself, we can do arbitrary pre-processing of the data before readying it for display. I have organized this sheet by putting labels for the data in column A and the values to be displayed in column B, but that’s incidental. In the next section we’ll see that data can come from any cell.
Template
An HTML template combines static HTML elements with special sentinels denoting variables to produce dynamic HTML output. I created an absolutely minimal templating language here, where template variables are surrounded by
{curly braces}, and they must refer to cells with data to display. There are plenty of fully-featured templating languages available – Handlebars is a famous one.As a small example, we could define a template like this:
<div> <i>Hello</i>, {B2}! </div>If the cell
B2in our sheet contained the textWorld, the resultant HTML would look like this:<div> <i>Hello</i>, World! </div>The part of the template that contained
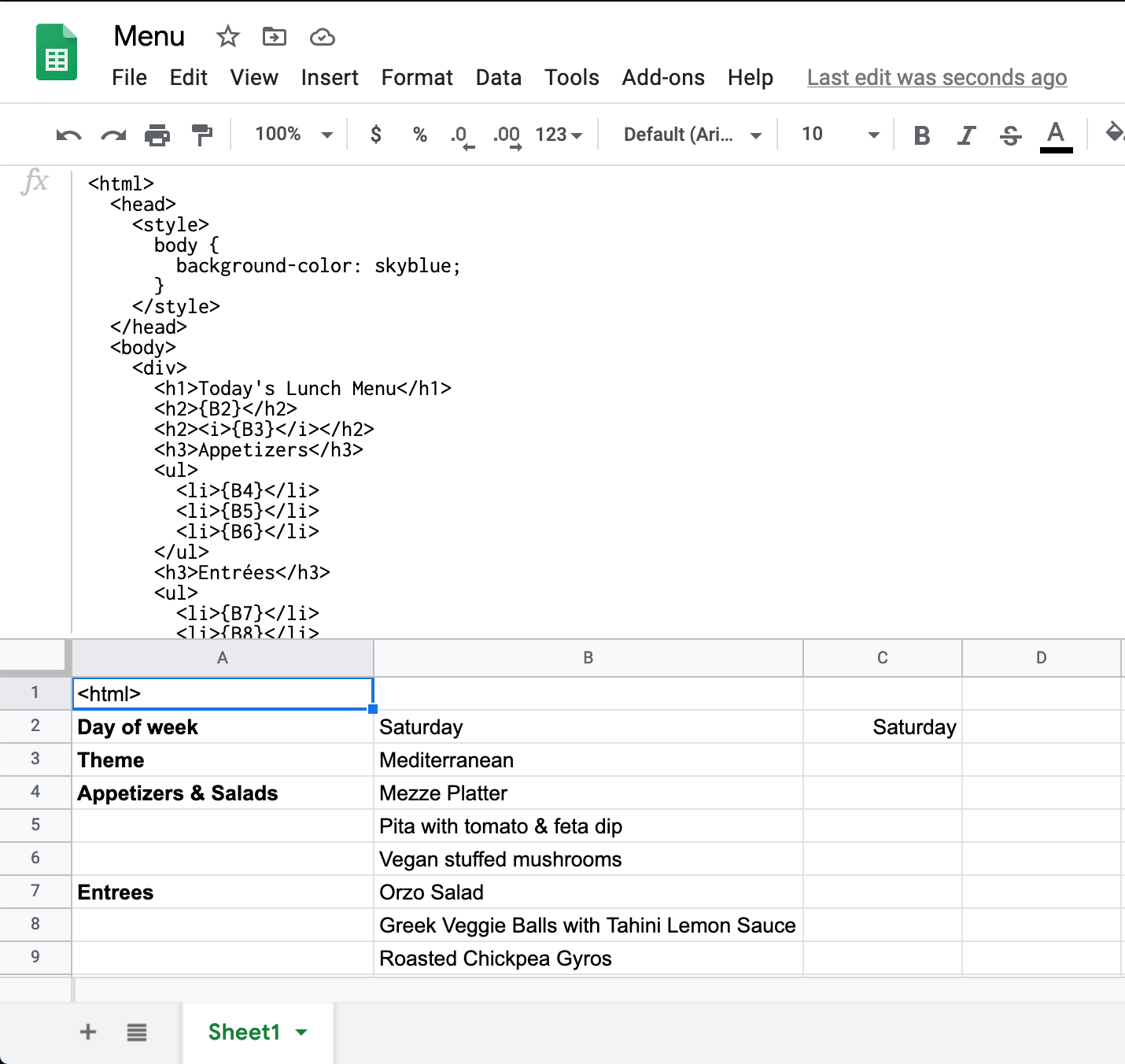
{B2}was replaced with the value from cellB2in our sheet – nice. So let’s build the HTML template for our menu, referencing the data cells defined in the previous section.<html> <head> <style> body { background-color: skyblue; } </style> </head> <body> <h1>Today's Lunch Menu</h1> <h2>{B2}</h2> <h2><i>{B3}</i></h2> <h3>Appetizers</h3> <ul> <li>{B4}</li> <li>{B5}</li> <li>{B6}</li> </ul> <h3>Entrées</h3> <ul> <li>{B7}</li> <li>{B8}</li> <li>{B9}</li> </ul> <p> Thanks for dining at the cafeteria! Please bus your table when you're finished eating! </p> </body> </html>This text should go in cell A1, which is where the script will read it from.
Script
Now, open the script editor by going to Tools > Script Editor. Here is the Apps Script script we’ll use to read data from the sheet, combine sheet variables with the HTML template, and serve that HTML in response to a GET request.
function letterToColumnOrdinal(letter) { // TODO: handle >= AA return letter.charCodeAt(0) - "A".charCodeAt(0); } function dataAtCellAddress(data, cellAddress) { const row = Number.parseInt(cellAddress.match(/\d+/)[0]) - 1; const col = letterToColumnOrdinal(cellAddress.match(/[A-Z]+/)[0]); return data[row][col]; } function doGet(e) { const sheet = SpreadsheetApp.getActiveSheet(); const data = sheet.getDataRange().getValues(); const template = data[0][0]; const html = template.replace(/{(.*?)}/g, function(match, p1) { return dataAtCellAddress(data, p1) }); return HtmlService.createHtmlOutput(html); }The
letterToColumnOrdinalfunction is just a helper function to take a letter from the cell address and convert it to a number so that we can index into thedatamatrix, e.g.C -> 2.dataAtCellAddresstakes the sheet data and a raw cell address and returns the value stored in that cell. ThedoGetfunction is called in response to aGETrequest. It is responsible for extracting data from the sheet and combining the cell data with the template string. When the site’s info or structure needs change, those modifications can happen directly in the sheet, and these 20 lines of Apps Script code won’t need to be touched.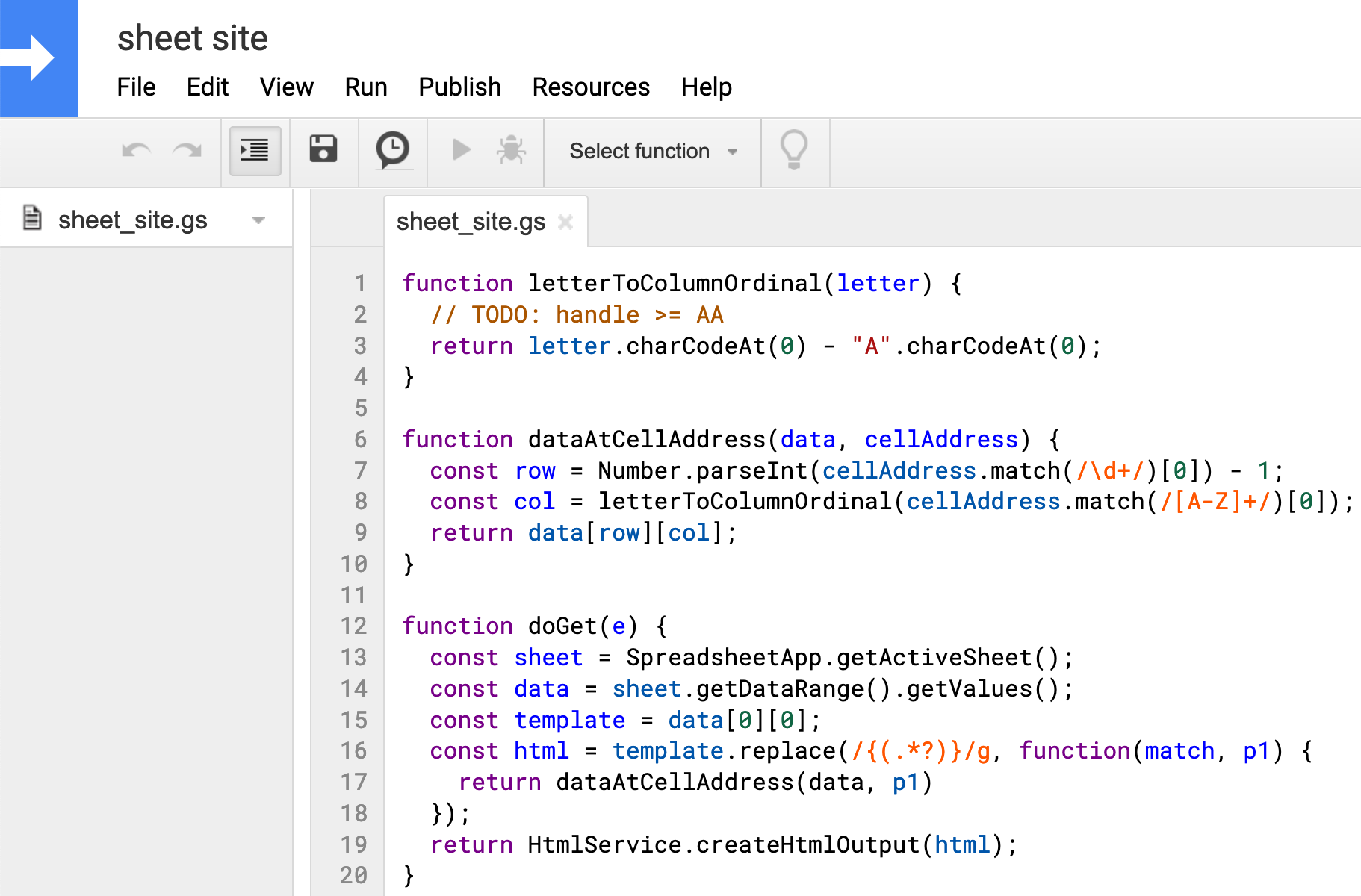
Deploy
Let’s deploy our new web page. From within the script editor, go to Publish > Deploy as web app.
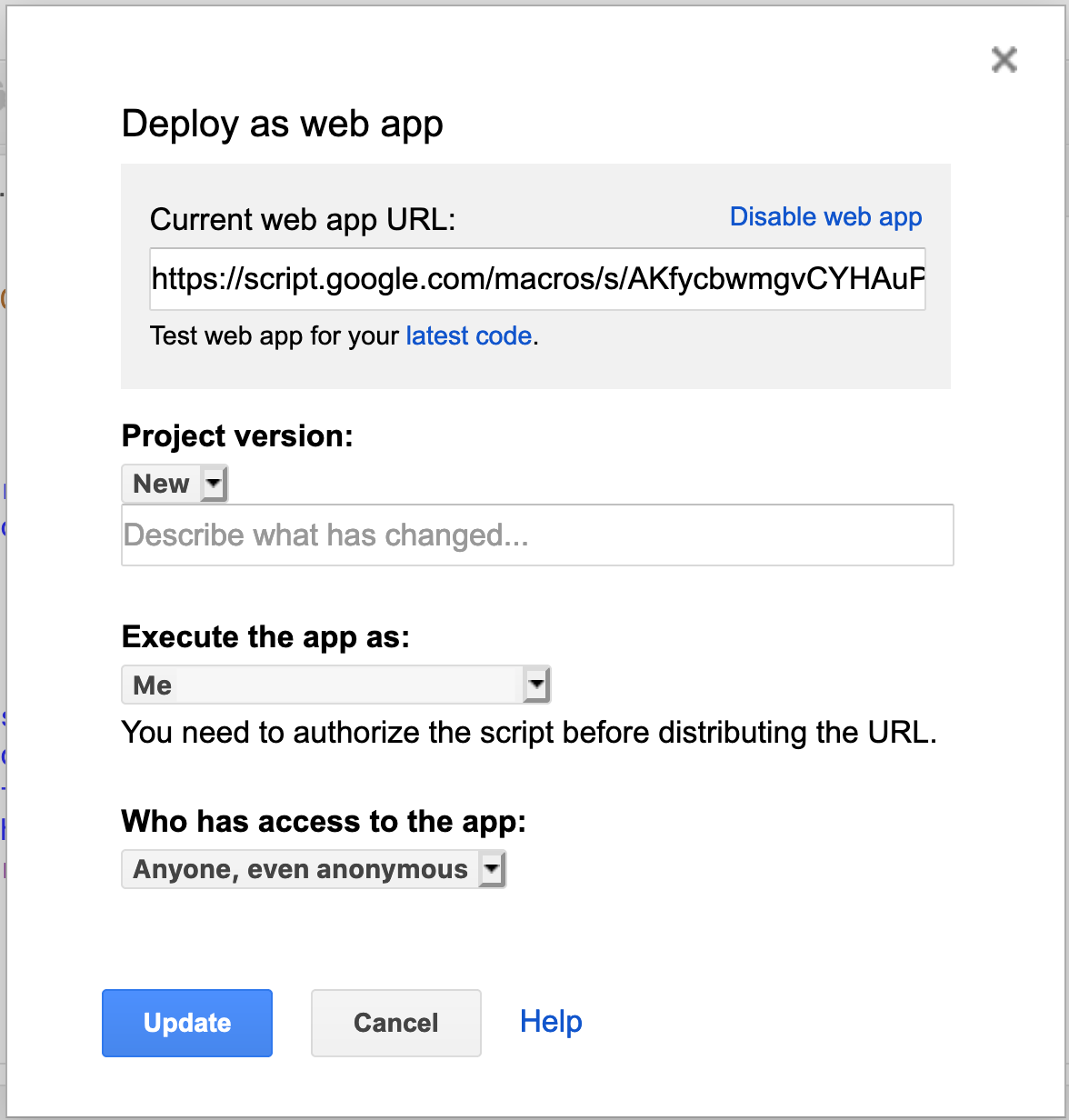
Then, fill out the dialog form and copy the URL to the web page. Here’s the example page I created: Lunch Menu.
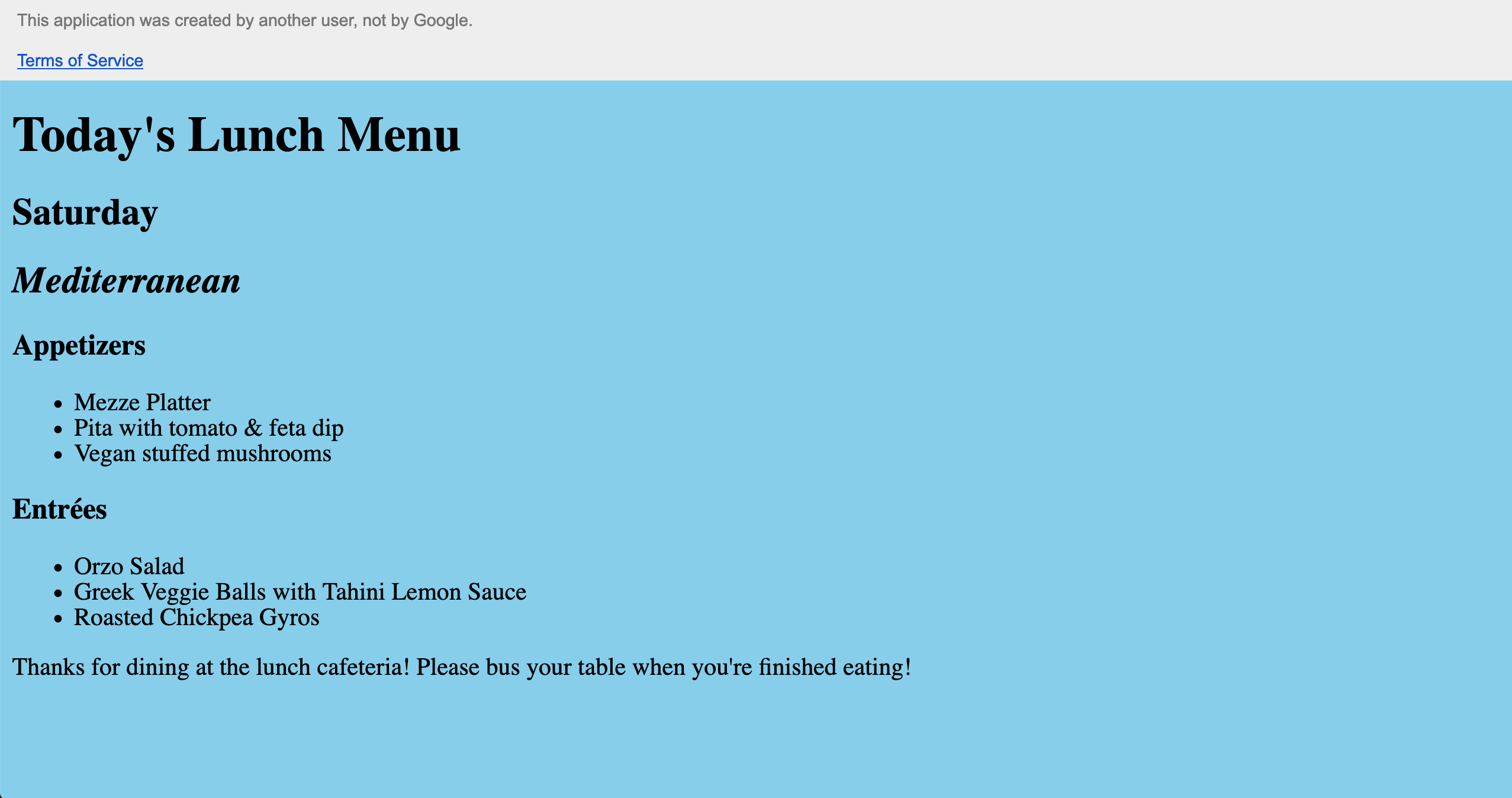
Great! Finally, let’s say the chef was inspired by this blog post to make a new app. So he’s eighty-sixing the mezze platter in favor of a bruschetta crostini. All we need to do is update that cell in the sheet…
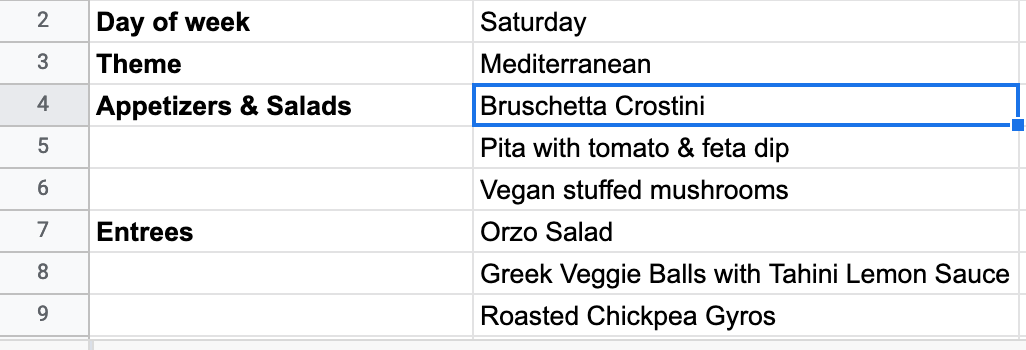
…and reload the web page.

And that’s it. We now have a simple, templatized web page that’s easy to change via the sheet.
-
Dec 31, 2019
A genetic algorithm Twitter art bot in Clojure
I recently released a project called
generation-p: an interactive art project driven by a genetic algorithm. The interface for this project is a Twitter bot. Once per day, the bot posts a new “individual” – a 32x32 pixel image – from the genetic algorithm’s population.This project explores the emergence of meaning from randomness. Can a population of images, which begins as noise, evolve into something worthy of attention? Retweets and favorites are a proxy for attention, and so they serve as inputs to the algorithm’s fitness function.
Over time, those images which garner more attention have a higher likelihood of being selected for reproduction, and the idea is that future generations will tend to produce more interesting images. In this way the bot is (very slowly) solving an optimization problem, where the target function is akin to attention-worthiness.
When I began work on this project, the output was textual rather than visual. I quickly pivoted to instead use images. Around early December I was looking at a lot of pixel art, and I loved how the artists were able to eek so much information (and evoke emotion) from so few colors and such little area. This project is partly inspired by pixel art, even if the results don’t look like much of anything. I think nascent patterns in images can be more quickly apparent, and reactions to them can be more immediate, and so images are well suited for this bot.
There are two crossover mechanisms at play. One method is k-point crossover, if the images were first flattened to one dimension. The other method is similar, but based on taking 2D regions from each of the parents. Also, the crossover methods and parameters are themselves inherited and undergo mutation. The selection mechanism is fitness proportional.
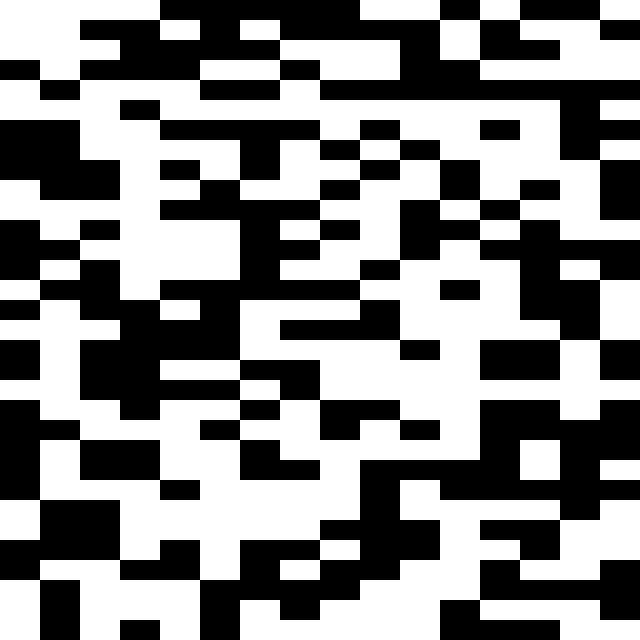
Below are examples of how the crossover methods control inheritance from either parent with different parameters.
Crossover with runs of length 2:
Length 8:

Patch crossover with patch-size 2:
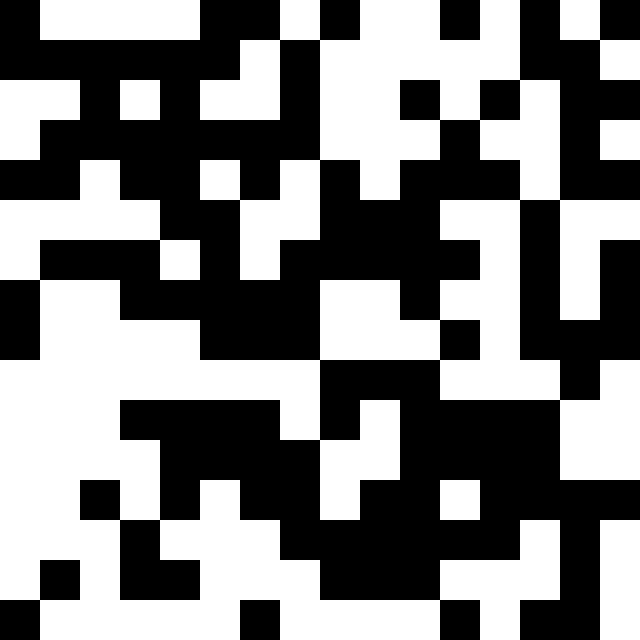
Patch-size 8:
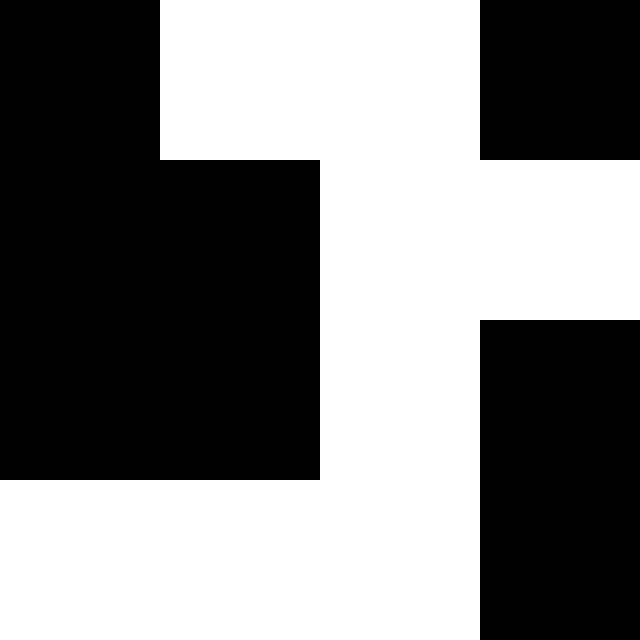
The number of individuals per generation, the size of the images, the mutation rates, and other inputs are all hard-coded hyperparameters that are pretty much arbitrary and mostly un-tuned. I suspect that whatever the output converges to will probably just look like noise, since the attention-worthiness fitness function is noisy. Even if everybody who interacted with the bot had a cohesive idea of what they wanted the output to look like, the continuous macaronage of crossover might prevent any “signal” in the image from emerging. But we shall see! I like the idea of a long-running bot like this where the output will change over months and years, because it mirrors the biological evolution we observe playing out in slow, animal time.
Writing this in Clojure was a joyful experience. I like REPL-driven development a lot, and it’s especially useful when experimenting with visual output. I found my crossover implementation to be sort of inelegant, but I think that’s just because my Clojure is rusty. Overall it was easy to get into a flow state with Clojure, and I’m really happy I got to use the language again after having not written anything in it for a long time.
-
Sep 28, 2019
Auto-saving Org Archive File
For note taking and TODO tracking, I use org mode for Emacs. When a TODO item is archived in an org buffer, it moves to a special archive file. This article is about how to automatically save the archive buffer after each item is archived.
When an item is archived, the archive file opens in a buffer, but the buffer isn’t saved. I find it annoying that I have to make special effort to save the archive file every time I archive something. This behavior could result in data loss if one neglects to save the archive file. Apparently “org-mode used to save that archive file after each archived item” [0]. I decided to re-implement that behavior in my own Emacs configuration.
Depending on your org setup, this can be accomplished relatively easily using
save-some-buffersandadvice-add.;; I have my primary org document's file name defined as a constant for use in ;; other parts of my config. (defconst notes-file "~/Documents/notes.org") ;; When an item is archived, it's saved to a file named ;; `<source-org-file>_archive`, so here I similarly define a constant for that ;; archive file. (defconst notes-archive-file (concat notes-file "_archive")) ;; The `save-some-buffers` function can run silently, and it also accepts a ;; function argument to predicate whether a buffer is saved or not. ;; This predicate simply says "save the buffer's file if it's the notes archive ;; file" (defun save-notes-archive-file () (interactive) (save-some-buffers 'no-confirm (lambda () (equal buffer-file-name (expand-file-name notes-archive-file))))) ;; Finally, the newly-defined function can advise the archive function. So, ;; after a subtree in org is archived, the archive file will be automatically saved. (advice-add 'org-archive-subtree :after #'save-notes-archive-file)References:
-
Feb 23, 2019
Noise generator
I often need to drown out distractions when trying to concentrate, and sometimes to do that I opt for using a noise generator. Searching for how to generate noise at the command line, I discovered this great answer on the Ubuntu Stack Exchange about using SoX.
Adapted for macOS (and assuming
brewis installed):brew install sox play -n synth brownnoise synth pinknoise mix synth sine amod 10 100I now have an alias to produce a pleasant mix of pink and Brown noise :D